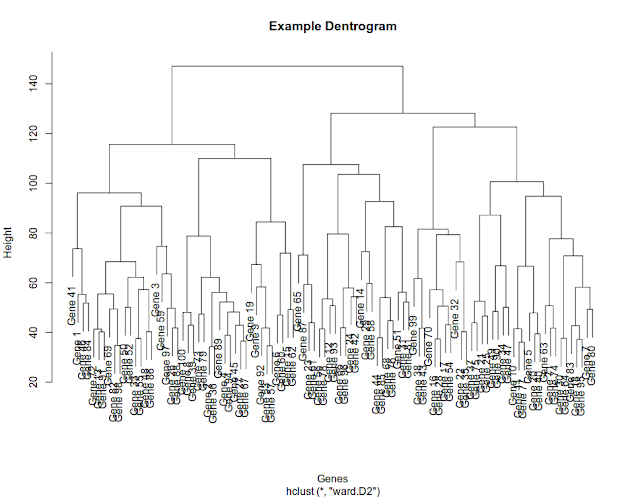
Many a times, very simple tasks seem to be difficult for eg. extracting members of a cluster. For a biologist, clustering of genes or pathways or samples very common. Then the question comes how to extract the genes of a particular cluster. Let us say you have a dendrogam with 7 clusters and you would like to extract genes of cluster 5.
Let us do a small simulation and then extract genes from a cluster of interest. Remember you can also do the same for any cluster or for any other variable (for eg sample, pathway, network module).
===================================Let us do a small simulation and then extract genes from a cluster of interest. Remember you can also do the same for any cluster or for any other variable (for eg sample, pathway, network module).
> data <- replicate(20, rnorm(100, mean =10, sd=10))
> rownames(data) <- paste("Gene", c(1:nrow(data)))
> colnames(data) <- paste("Sample", c(1:ncol(data)))
> library(stats)
> d <- dist(data, method = "euclidean")
> hc <- hclust(d, method = "ward.D2")
> plot(hc)
Now how do we extract the genes of interest? To do that, first we need to define the number of clusters in such a way that genes of interest fall in an independent cluster. How do we do that? We keep partitioning dendrogram till we identify the cluster with genes of interest. We can partition the data by defining a limit/boundary on height of dendrogram. For eg. in above diagram, if we cut the tree at 130 , we are left with three clusters (count always from the top, highlighted in red color below)
Now let us say, we cut the tree at 110, we will have 5 clusters.
Now we know how to partition the dendrogram. After partitioning the dendrogram, we can extract the genes from the cluster of interest. For eg. let us say we want to extract genes from 5th cluster after partitioning the data with a height of 110.
Let us extract all the genes for which cluster is assigned 5 and their expression values.
============================================== # Cut the tree at 110
h=110
k=cutree(hc,h=h)
# Extract the genes assigned to cluster 5
names(k[k==5])
# Now extract the expression values for the genes in cluster 5
data[names(k[k==5]),]
===============================================
(Note: Dendrogram produced above may not be same each time we run the code as data is randomly simulated. Set seed to get consistent dendrograms)