libraries required: Biostrings (for later purpose)
Create random nucleotide sequence of length 1000 bases:
1) Load library Biostringscode: library(Biostrings)
2) Generate random sequence of length 1000 bases with letters ATGC only. This would print 1000 base length nucleotide characters.
code: sample(DNA_BASES, 1000, replace = TRUE, prob = c(0.4,0.1,0.1,0.4))
Function DNA_BASES in biostrings prints 4 bases, where DNA_ALPHABETS prints entire IUPAC code for DNA.
Please note that in sample function, we can supply probability as well, for each base. In the example above, we have supplied example probabilities for each base.
3) Save the output as an object in R.
code: seq.n=sample(DNA_BASES, 1000, replace = TRUE, prob = c(0.4,0.1,0.1,0.4))
4) Collapse all characters in to a single sequence
command: paste(seq.n, collapse="")
This would print a continuous/contiguous stretch of bases (4). However, we would like to use it for further processes. At this point this can either be saved as another object or as a biostring.
5) Let us save it as biostring object.
command: ds.seq.n=DNAString(paste(seq.n, collapse=""))
6) Look at the object class.
commad: class (ds.seq.n)
This would print the class as biostring.
7) Let us look at the frequency of the bases. This should match with the probabilities supplied in 2 and 3rd steps
command: alphabetFrequency(ds.seq.n, baseOnly=TRUE)
Please note that baseOnly option prints frequencies for A, T, G, C and others. If this options is not supplied, frequencies for all IUPAC bases will be printed.
8) Write the file to disk.
First we need to convert the DNAstring to DNAstringset as DNAstring cannot be directly written to disk in fasta format. In addition, we cannot supply name(s) to DNAstring object where as DNAstringset object can accept names. Please follow the code for comments:
Code:
dss.seq.n=DNAStringSet(ds.seq.n) # converts and stores DNAstring object as DNAstringSet object
names(dss.seq.n)="test.nt" # name of the sequence is set to test.nt
writeXStringSet(dss.seq.n, "test.fasta", format = 'fasta') # writes the above example random file to current working directory (in R) as test.fasta.
9) Open the file (test.fasta) in any text editor of your choice.
Code for generating random nucleotide sequence:
library(Biostrings)
seq.n=sample(DNA_BASES, 1000, replace = TRUE, prob = c(0.4,0.1,0.1,0.4))
ds.seq.n=DNAString(paste(seq.n, collapse=""))
class(ds.seq.n)
alphabetFrequency(ds.seq.n, baseOnly=TRUE)
dss.seq.n=DNAStringSet(ds.seq.n)
names(dss.seq.n)="test.nt"
writeXStringSet(dss.seq.n, "test.fasta", format = 'fasta')
Create random aminoacid sequence of length 350 amino acids:
1) Load library Biostringscode: library(Biostrings)
2) Generate random sequence of length 350 amino acids (AA) with standard 20 aminoacids. This would print 350 amino acids length characters.
code: sample(c(AA_STANDARD), 350, replace = TRUE)
Function AA_STANDARD in biostrings prints 20 AA, where AA_ALPHABET prints entire IUPAC code for proteins. Please note that in sample function, we can supply probability as well, for each AA. However, In the example above, we have not supplied example probabilities for each AA.
3) Save the output as an object in R.
code: seq.p=sample(AA_STANDARD, 350, replace = TRUE)
4) Collapse all characters in to a single sequence
command: paste(seq.p, collapse="")
This would print a continuous/contiguous stretch of AA. However, we would like to use it for further processes. At this point this can either be saved as another object or as a biostring.
5) Let us save it as biostring object.
command: ds.seq.p=AAString(paste(seq.p, collapse=""))
Please note that biostring objects for Nt (DNAstring) and AA (AAstring) are different as biostring looks at the standard code for each type.
6) Look at the object class.
command: class (ds.seq.p)
This would print the class as biostring.
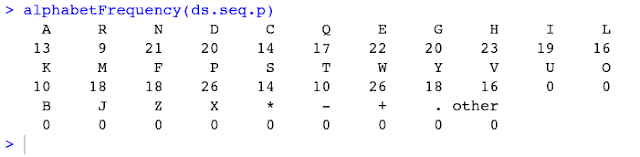
7) Let us look at the frequency of AA.
command: alphabetFrequency(ds.seq.p)
8) Write the file to disk.
First we need to convert the AAstring to AAstringset as AAstring cannot be directly written to disk in fasta format. In addition, we cannot supply name(s) to AAstring object where as AAstringset object can accept names. Please follow the code for comments:
Code:
dss.seq.p=AAStringSet(ds.seq.p) # converts and stores AAstring object as AAstringSet object
names(dss.seq.p)= "test.p" # name of the sequence is set to test.p
writeXStringSet(dss.seq.p, "testp.fasta", format = 'fasta') # writes the above example random AA file to current working directory (in R) as testp.fasta.
9) Open the file (testp.fasta) in any text editor of your choice.
Code for creating random amino acid sequence:
seq.p=sample(AA_STANDARD, 350, replace = TRUE)
seq.p
ds.seq.p=AAString(paste(seq.p, collapse=""))
ds.seq.p
alphabetFrequency(ds.seq.p)
# write file to disk
dss.seq.p=AAStringSet(ds.seq.p)
names(dss.seq.p)= "test.p"
writeXStringSet(dss.seq.p, "testp.fasta", format = 'fasta')