In general, genome coordinates in VCF and reference fasta files will be in either hgxx (for eg hg19, hg38) or b3x (b32, b35)
format.
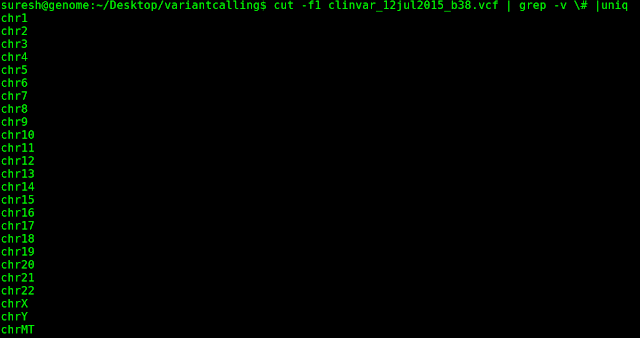
Before:
After:
hgxx format will have "chr" prefix to each chromosome number and Mitochondrial (chrM) chromosome will be at the start. b3x format will have no prefix for chromosome numbers and Mitochondrial chromosome will be at the last. If one has used hg19 reference (fasta from UCSC) in variant calling ( and prior steps in NGS analysis) and would like to annotate using clinvar/dbSNP, user has to append string "chr" to the first column "#CHROM" in VCF. To append string "chr" to chromosome numbers in #CHROM column, use the following command:
command:
sed 's/^\([1-9,X,Y,MT]\)/chr&/g' clinvar_GRCh38_12jul2015.vcf > clinvar_12jul2015_b38.vcf
One can download clinvar vcf file from ftp://ftp.ncbi.nlm.nih.gov/snp/organisms/human_9606_b142_GRCh38/VCF/ for GRCh 38.
Before:
After: