Some times, we may need ideogram information for the organism of our interest. Let us assume that UCSC (genome browser) supports that. To export ideogram information from UCSC, follow the instructions given below. Please note that the ideogram information can be exported as text (displayed within your browser of choice) or as gzipped file.
1) Navigate to table browser page hosted on UCSC
URL: https://genome.ucsc.edu/cgi-bin/hgTables
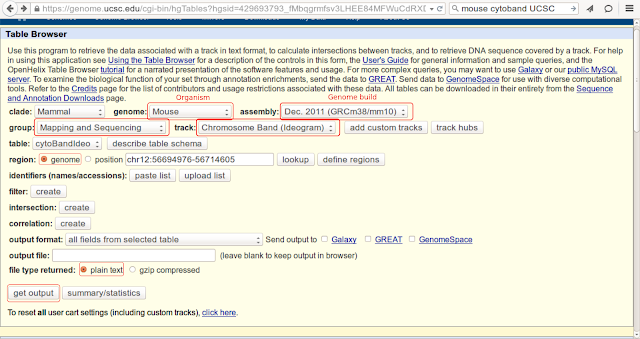
2) Page would look like this:
3) Select following fields to export ideogram information (from drop down menu for each filed):
1) Navigate to table browser page hosted on UCSC
URL: https://genome.ucsc.edu/cgi-bin/hgTables
2) Page would look like this:
3) Select following fields to export ideogram information (from drop down menu for each filed):
- Genome: Organism of interest (for eg. mouse)
- Assembly: Genome assembly for that organism (for eg. GRCm38/mm10) for mouse
- Group: Mapping and Sequencing
- Track: Chromosome Band (Ideogram)
- Region: choose radio button again genome (by default it would stay selected)
- At the end, select one of the formats you would like to download by selecting against field "file type returned".
- Click on "get output" button.
- If you select, "plain text", your results would be displayed in the browser it self which can be saved as text. If you choose "gzip compressed", you are asked download gzipped file. Download the file and gunzip it in linux or in windows, any modern unzip utilities will be able to gunzip the file.