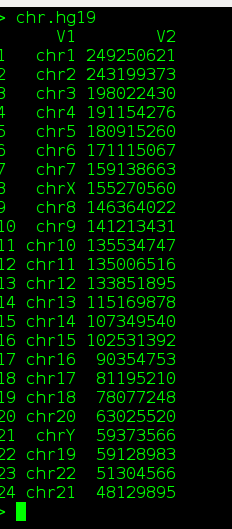
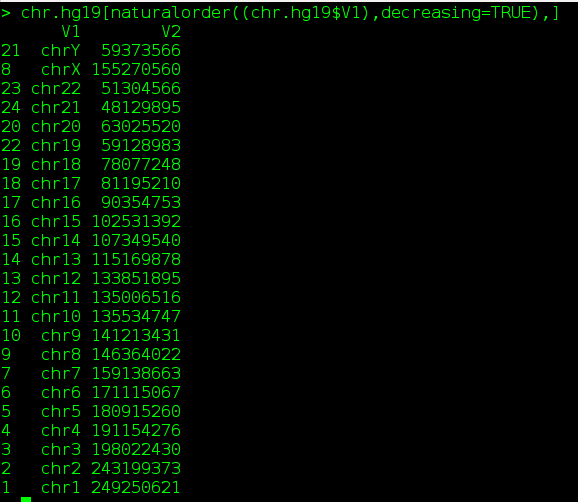
Some times, trivial issues bother. Here is a small tip to sort a chromosomes (values in a column) in human readable chronological order. This sorting is also called alpha, numeric sort. Following note, gives a general example to sort a data frame in natural order based on any user mentioned column.
Requirements:
package "naturalsort" in R
Commands:
1. data.frame[naturalorder(vector in dataframe), ] (by default, the values are sorted ascending order)
Note: This would sort the data frame in natural sort as per the values in mentioned vector in data frame, in ascending order.
2. data.frame[naturalorder((vector in dataframe), decreasing=TRUE), ]
(by default, the values are sorted ascending order)
Note: This would sort the data frame in natural sort as per the values in metioned vector in data frame, in descending order.
Example:
1. chr.hg19[naturalorder(chr.hg19$V1),]
This would sort chrosomes from chromosome 1 to chromsome XY.
2. chr.hg19[naturalorder((chr.hg19$V1),decreasing=TRUE),]
This would sort chrosomes from chromosome XY to chromsome 1.
Note: chr.hg19 is data frame created from http://genome.ucsc.edu/goldenpath/help/hg19.chrom.sizes and taking only chromosomes.
Case study:
1) Import chromosome sizes from UCSC
2) Sort the chromosomes in human readable order.
Method:
1) Load library "naturalsort" in R.
2) Download the data and store it as object in R.
command: chr.hg19=read.delim(url("http://genome.ucsc.edu/goldenpath/help/hg19.chrom.sizes"),
3) Print chr.hg19
command: dim(chr.hg19)
This would display tha there are 93 rows and with 2 columns. First colum contains chromosomes, other artefacts. Column two contains lengths of the chromosomes in columns in bp.
5) Now, for some reason, if you need to sort in reverse order (ie. descending fashion), following is the command:
command: chr.hg19[naturalorder((chr.hg19$V1),decreasing=TRUE),]
All commands:
library(naturalsort) ## loads the library naturalsort
chr.hg19=read.delim(url("http://genome.ucsc.edu/goldenpath/help/hg19.chrom.sizes"), header=FALSE) ## downloads the chromomosome sizes and stores it as object "chr.hg19"
chr.hg19=chr.hg19[1:24,] # extracts first 24 lines (chromosomes)
chr.hg19.asc=chr.hg19[naturalorder(chr.hg19$V1),] ## sorts the chromosomes in natural order in ascending fashion
chr.hg19.desc=chr.hg19[naturalorder((chr.hg19$V1), decreasing=TRUE),] ## sorts the chromosomes in natural order in descending fashion.
Requirements:
package "naturalsort" in R
Commands:
1. data.frame[naturalorder(vector in dataframe), ] (by default, the values are sorted ascending order)
Note: This would sort the data frame in natural sort as per the values in mentioned vector in data frame, in ascending order.
2. data.frame[naturalorder((vector in dataframe), decreasing=TRUE), ]
(by default, the values are sorted ascending order)
Note: This would sort the data frame in natural sort as per the values in metioned vector in data frame, in descending order.
Example:
1. chr.hg19[naturalorder(chr.hg19$V1),]
This would sort chrosomes from chromosome 1 to chromsome XY.
2. chr.hg19[naturalorder((chr.hg19$V1),decreasing=TRUE),]
This would sort chrosomes from chromosome XY to chromsome 1.
Note: chr.hg19 is data frame created from http://genome.ucsc.edu/goldenpath/help/hg19.chrom.sizes and taking only chromosomes.
Case study:
1) Import chromosome sizes from UCSC
2) Sort the chromosomes in human readable order.
Method:
1) Load library "naturalsort" in R.
2) Download the data and store it as object in R.
command: chr.hg19=read.delim(url("http://genome.ucsc.edu/goldenpath/help/hg19.chrom.sizes"),
3) Print chr.hg19
command: dim(chr.hg19)
This would display tha there are 93 rows and with 2 columns. First colum contains chromosomes, other artefacts. Column two contains lengths of the chromosomes in columns in bp.
3) Since we deal with chromosomes only, we need to extract only chromosomes and their coordinates. We know that chromosomes are 24 (22 autosomes + XY).
Command: chr29=chr.hg19[1:24,]
This would save first 24 entries/chromosomes as a data.frame. Please note that chromosomes are sorted numerically (column V1).
4) Sort chromosomes in human/alpha numeric order, in descending order i.e from chr1 to chrx and y.
command: chr.hg19.asc=chr.hg19[naturalorder(chr.hg19$V1),]5) Now, for some reason, if you need to sort in reverse order (ie. descending fashion), following is the command:
command: chr.hg19[naturalorder((chr.hg19$V1),decreasing=TRUE),]
All commands:
library(naturalsort) ## loads the library naturalsort
chr.hg19=read.delim(url("http://genome.ucsc.edu/goldenpath/help/hg19.chrom.sizes"), header=FALSE) ## downloads the chromomosome sizes and stores it as object "chr.hg19"
chr.hg19=chr.hg19[1:24,] # extracts first 24 lines (chromosomes)
chr.hg19.asc=chr.hg19[naturalorder(chr.hg19$V1),] ## sorts the chromosomes in natural order in ascending fashion
chr.hg19.desc=chr.hg19[naturalorder((chr.hg19$V1), decreasing=TRUE),] ## sorts the chromosomes in natural order in descending fashion.