Some times, user may have to replace all the values with some other values for eg. all the gene symbols with entrez gene IDs. Now let us have a look at following data and data can be downloaded from here:
$ test$ncbi_gene_id=as.character(test$ncbi_gene_id)
$ library(forcats)
$ new_df=df
$ new_df[-1] <- lapply(new_df[-1], function(x) lvls_revalue(factor(x, levels = test$hgnc_symbol), test$ncbi_gene_id))
===========================================
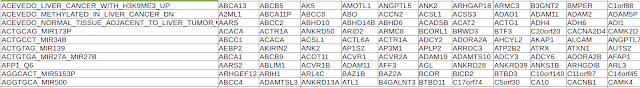
Let us have a look at new_df (output) now:
This data has IDs in first column and rest of the columns with HGNC symbols. Now we need to replace all of them with entrez gene IDs. Let us do it with forcats package.
Steps are to make a unique list of genes and get Entrez IDs using biomart library. Then map them to gene symbols and then replace.
=========================================
$ library(tidyr)
$ gene_list=unique(sort(gather(df[-1],"","genes")$genes))
$ gene_list=unique(sort(gather(df[-1],"","genes")$genes))
$ library(biomaRt)
$ mart <- useMart(biomart = "ensembl", dataset = "hsapiens_gene_ensembl")
$ test=getBM(attributes = c("hgnc_symbol", "entrezgene"), filters = "hgnc_symbol", values = gene_list, bmHeader = T, mart = mart)
$ colnames(test)=c("hgnc_symbol","ncbi_gene_id")$ mart <- useMart(biomart = "ensembl", dataset = "hsapiens_gene_ensembl")
$ test=getBM(attributes = c("hgnc_symbol", "entrezgene"), filters = "hgnc_symbol", values = gene_list, bmHeader = T, mart = mart)
$ test$ncbi_gene_id=as.character(test$ncbi_gene_id)
$ library(forcats)
$ new_df=df
$ new_df[-1] <- lapply(new_df[-1], function(x) lvls_revalue(factor(x, levels = test$hgnc_symbol), test$ncbi_gene_id))
===========================================
Let us have a look at new_df (output) now: