Here is a simple row wise function. Following are assumptions:
- There are two conditions
- Column numbers are mutually exclusive and full (for eg if 4 columns are one group, rest 4 columns are second group for a 8 column matrix with row names as gene names, sample names as column names)
- Function doesn't have any other options that are offered by t.test. For advanced t.test please use t.test function it self with all the options.
- This function is a very simple script for row wise t test.
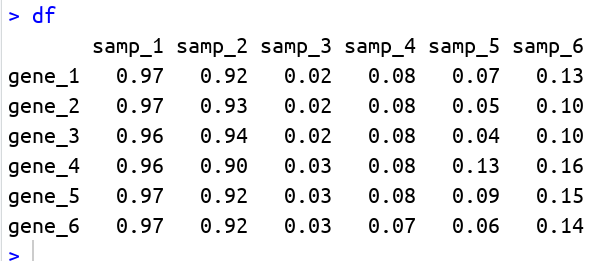
matrix is follows:
As you see above matrix has 6 samples and 6 genes. Number of genes and number of samples arbitrary here. So we are supposed to do row wise t tests with first three samples (samp_1 to samp_3) as one group, rest samples are in other group. t-test result in R stores the p-value in p.value and let us extract that and append it to the input matrix. Let us write the function and run the program.
========================================================
Ttestpalue <- function(df, col1, col2) {
df$pvalue<-apply(df,1, function(x) t.test(x[as.integer(col1):as.integer(col2)],x[as.integer(col2)+1: length(x)], var.equal = F)$p.value)
return(df)
}
df$pvalue<-apply(df,1, function(x) t.test(x[as.integer(col1):as.integer(col2)],x[as.integer(col2)+1: length(x)], var.equal = F)$p.value)
return(df)
}
======================================================-
Input for the function is matrix, column numbers are starting sample column and end column of the samples belong to one group. Rest samples are automatically grouped to another group.
- Let us load the dummy data and you can download it here.Rename the downloaded file to "test.txt"
Data=read.csv("test.txt", header = T, stringsAsFactors = F, strip.white = T, sep="\t", row.names = 1) - Execute above function and a new function is created
- Execute following code to get the row wise p value.
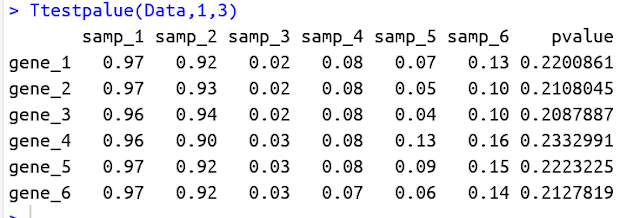
Ttestpalue(Data,1,3) - Now the output would look like this: