For basic processing of the text, AWK and Sed are two of the best CLI tools available in GNU-linux. Today, let us look at conditional filtering of columns using a text file and download it here.
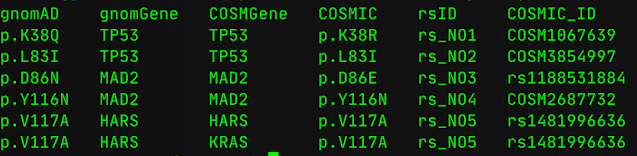
Text file looks as below:
Now, one has to compare if Gene from 2nd column (gnomGene) matches with 3rd column (COSMGene) and if it matches, compare column 1 (variants in gnomAD column) and 4 (variants in COSMIC column). If column 1 matches with column 4, then put in a "match" column and if column 1 variant doesn't match column 4 variants, put in a "Nomatch" column.
code:
==========================================
$ awk -v OFS="\t" -F"\t" 'NR==1 {print $0,"Match","NoMatch"}; NR>1 {if($2==$3){if($1==$4) {$7=$1} else {$8=$1}} else {};print}' file.txt
==========================================
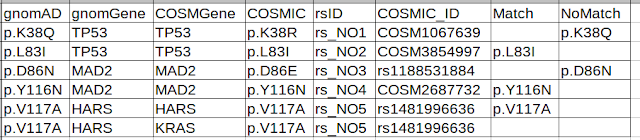
Final output looks like this:
Explanation of the code:
- NR==1 {print $0,"Match","NoMatch"} -- prints header of the original file and two new columns names "Match" and "NoMatch"
- NR>1 {if($2==$3){if($1==$4) {$7=$1} else {$8=$1}} else {};print} -- a nested if..else condition. First if checks column 2 matches with column3. If (first) matches, then second if kicks in. Second if checks, if first column is equivalent to 4th column. If it is equivalent, then column 1 value will be put in column 7 (match). If it is not equivalent, then column 1 value will be put in column 8 (nomatch). If first if fails (column 2 doesn't match with column 3), no action happens i.e both match and nomatch columns will be left empty.
- print -- prints the file at the end